Diet, Obesity, And
Type 2 Diabetes
pROJECT dETAILS
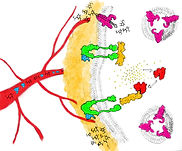
An illustration depicting the relationship between high-fat diets, obesity, and insulin resistance in Type 2 Diabetes. The purpose of the project was to illustrate a scientifically accurate molecular interaction for the educated layperson, in the form of a Scientific American style figure. Protein models were obtained from the Protein Data Bank and refined in Chimera for use in Cinema 4D. The final illustration was arranged in Adobe Illustrator.
Audience
General
Client
Dr. Derek Ng
Completed
July 2018
Medium
Chimera
Maxon Cinema 4D
Adobe Photoshop
Adobe Illustrator
FORMAT
11 x 17 (print)



ProCESS Work
Initial Research
The challenge for this project was to create a visualization that depicted the structure-function relationship of a dynamic molecular process. Early on I decided I wanted to focus on a topic related to diabetes, as having Type 1 diabetes myself, the molecular mechanisms of the disease have always interested me, as have the differences between Type 1 and Type 2 diabetes. My initial research into potential topics involved narrowing down an appropriate molecular pathway and conducting a media audit of visualizations that depicted similar processes. In completing this initial research, I determined that Type 2 diabetes, and specifically the role diet and obesity play in developing insulin resistance, offered an interesting molecular process that was well documented in the literature—a necessity for creating an accurate visualization.

THUMBNAILS & REFINED RESEARCH
Once I had decided on the molecular mechanism I wanted to visualize, I needed to determine the best way to illustrate the process and characters involved. As I intended to show how a high-fat diet and obesity could lead to insulin resistance and type 2 diabetes, I wanted to contrast the normal and pathological pathways and drew out several thumbnails of varying complexity which compared the two. Some of these initial designs offered a more traditional, simplistic comparative approach; however, I wanted to attempt a more complicated depiction that placed the audience inside of the molecular environment and dynamically guided them through the process. Completing the thumbnails also allowed me to determine who my characters were for the scene—that is the proteins and structures that needed to be visualized and rendered—as well as the environment they should be placed in. I created a reference list of these characters and environments to guide my design moving forward, which outlined their appearance, motion, interactions, and linked to their Protein Data Bank (PBD) structural data files if available.





DETERMINING THE NARRATIVE COMPOSITION
After my thumbnails and character reference sheet were created, the next step was to experiment with the arrangement of my visual and text elements to best facilitate the narrative I wanted to communicate. I decided to combine several different elements of my thumbnails in order to create a story that introduced the audience to the topic and took them through the gross-anatomical pathway they were likely most familiar with before guiding them through the more complex molecular processes. At this point, the composition and written content were essentially finalized, meaning I could complete a near-final composite sketch and confidently move to the production stage of the project.



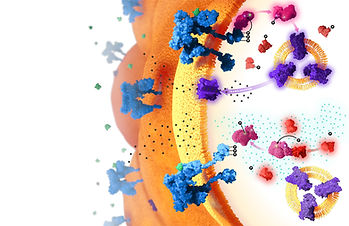
Creating an Accurate Molecular Environment
While the most technically complex part of the project, the bulk of the work necessary to create a believable and accurate molecular environment was completed prior to the production phase. Using the character reference sheet made during my initial research, I was able to refer to and import protein models from PBD into Chimera, a program designed for the visualization and analysis of molecular structures. Once in Chimera, I could import the protein structures as OBJ files into Cinema 4D (C4D), the 3D modelling and rendering tool used in this project. I first created the molecular environment the proteins needed to exist in, using an online tutorial to model a lipid bilayer. As the bilayer itself was somewhat resource intensive to render and visualize, I used a combination of depth-of-field and a noise texture to approximate the surface of adipose tissue receding into the background. Next, I added each molecular character to the scene, adjusting their position, texture, and the overall lighting as I progressed.



Final renders & Arrangement
At this point in the project, the final arrangement was compiled in Adobe Illustrator using exported assets from C4D (for in the molecular environment) and Photoshop (for the anatomical illustrations). The molecular structures from C4D were rendered and exported as separate files, meaning they maintained the appearance of existing in the same scene, but they could be individually moved in Illustrator to facilitate any necessary adjustments for the final composition. Similarly, small tweaks were made to the text arraignment and direction of the anatomical illustration to better guide the audience through the piece. The final composition was exported from Adobe Illustrator with the intention of existing as an 11x17 inch magazine spread.



References
Khodabandehloo H, Gorgani-Firuzjaee S, Panahi G, Meshkani R. Molecular and cellular mechanisms linking inflammation to insulin resistance and β-cell dysfunction. Transl Res. 2016;167(1):228-256. doi:10.1016/j.trsl.2015.08.011; Lizcano JM, Alessi DR. The insulin signalling pathway. Curr Biol. 2002;12(7):236-238. doi:10.1016/S0960-9822(02)00777-7.; Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414(6865):799-806. doi:10.1038/414799a.; Gustafson B, Hedjazifar S, Gogg S, Hammarstedt A, Smith U. Insulin resistance and impaired adipogenesis. Trends Endocrinol Metab. 2015;26(4):193-200. doi:10.1016/j.tem.2015.01.006.; Mohan SS, Perry JJP, Poulose N, Nair BG, Anilkumar G. Homology modeling of glut4, an insulin regulated facilitated glucose transporter and docking studies with atp and its inhibitors. J Biomol Struct Dyn. 2009;26(4):455-464. doi:10.1080/07391102.2009.10507260; Kamata H, Honda SI, Maeda S, Chang L, Hirata H, Karin M. Reactive oxygen species promote TNFα-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120(5):649-661. doi:10.1016/j.cell.2004.12.041; Guo H, Callaway JB, Ting JPY. Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21(7):677-687. doi:10.1038/nm.3893.